Chinese Scholars Make New Progress in Genome Decoding
Supported by the National Natural Science Foundation of China (Grant Nos. 32588201, 31925015, 32161133016, 323B2011), Professor Shen Xiaohua’s team at Tsinghua University has achieved new progress in genome decoding. The researchers developed a high-throughput single-cell nascent transcriptome technology (scFLUENT-seq), which, for the first time, quantifies genome usage at the single-cell level. The study reveals the sparsity and heterogeneity of nascent transcription, offering fresh insights into the diversity of cell fates. The work, entitled Single-cell nascent transcription reveals sparse genome usage and plasticity, was published online in Cell on September 26, 2025 (DOI: https://doi.org/10.1016/j.cell.2025.09.003).
The genome serves as life’s blueprint, yet how much of it is actually used by a single cell has remained unclear. Shen’s team developed single-cell full-length EU-labeling and the high-throughput 10X genomics platform. They discovered that even in highly transcriptionally active mouse embryonic stem cells, only about 3% of the genome is transcribed at any given time—though this is more than 50-fold higher than in terminally differentiated splenic lymphocytes. When data from all cells are combined, nascent transcription covers nearly 80% of the genome. This striking contrast underscores the extreme sparsity, randomness, and heterogeneity of transcription at the single-cell level—patterns that are largely obscured in bulk analyses.
The study also shows that transcription in noncoding regions is more stochastic than in protein-coding genes. Distal noncoding transcription units in heterochromatic regions, although sparse, exhibit regulatory properties shaped by their chromatin environment. The team introduced the concept of “nascent transcriptional diversity” to describe transient and unstable cellular states. Such cells, indistinguishable in steady-state transcriptomes, simultaneously transcribe more protein-coding genes and noncoding units at the nascent level, reflecting heightened fate plasticity.
This research not only establishes precise single-cell measurement of nascent transcription but also addresses two central theoretical questions: the transcriptional activity of noncoding “dark matter” and the inherently probabilistic nature of life regulation. By quantitatively uncovering the sparsity and heterogeneity of nascent transcription, the study moves beyond the limitations of steady-state transcriptomics and provides a new framework for rethinking genome regulation and cell fate.
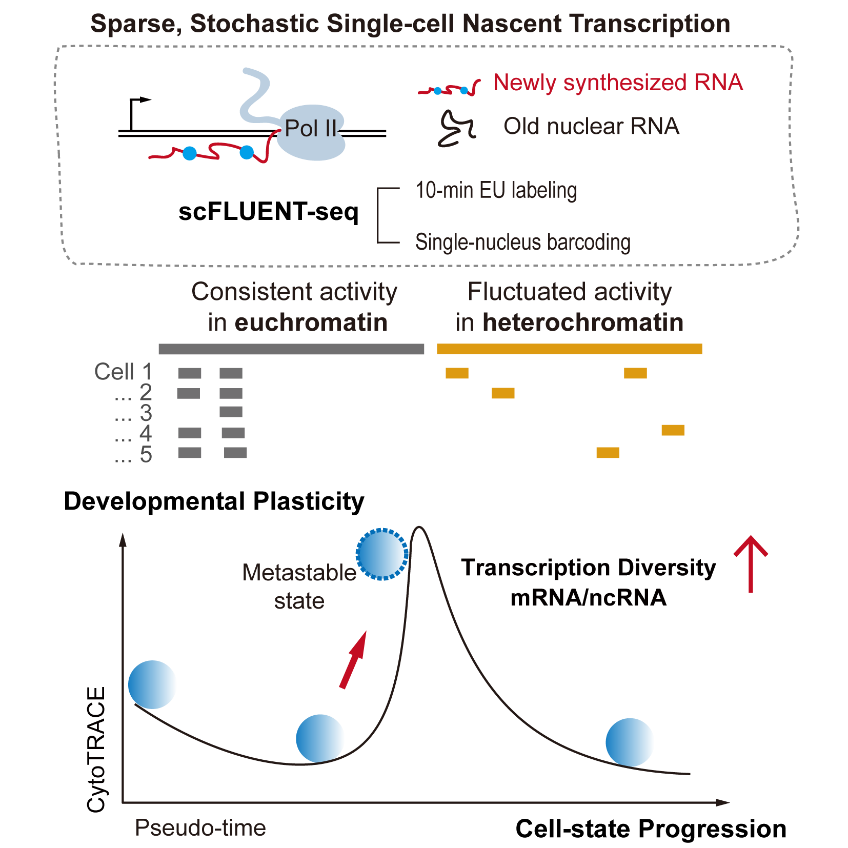
Figure. Single-cell nascent transcriptome profiling and transcriptional heterogeneity and plasticity
Contact Us

National Natural Science Foundation of China
Add: 83 Shuangqing Rd., Haidian District, Beijing, China
Postcode: 100085
Tel: 86-10-62327001
Fax: 86-10-62327004
E-mail: bic@nsfc.gov.cn
